Using Kydavra KullbackLeiblerSelector for feature selection

What is KullbackLeiblerSelector ?
It is a feature selector based on the Kullback-Leibler divergence.
A divergence is a measure of difference between two probabilistic distributions. In the case of machine learning, we can consider data distributions and calculate how different a certain feature column is compared to a target column.
How is it calculated ?
Kullback-Leibler divergence, also named relative entropy in information theory, is calculated using the following formula:
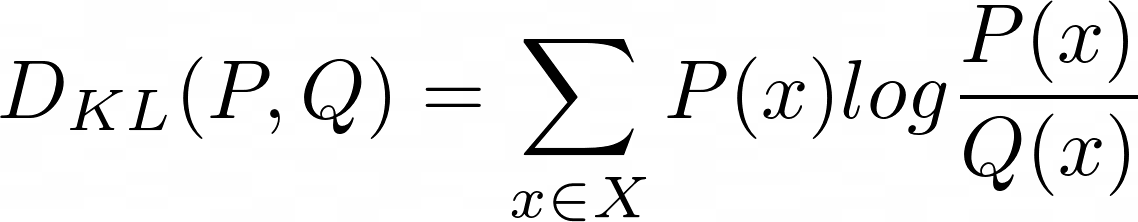
where P and Q are the distributions and X is the sample space. This is for discrete distributions.
Using KullbackLeiblerSelector
The KullbackLeiblerSelector constructor has the following parameters:
- EPS (float, default: 0.0001): a small value to add to the feature column in order to avoid division by zero.
- min_divergence (int, default: 0): the minimum accepted divergence with the target column
- max_divergence (int, default: 1): the maximum accepted divergence with the target column
The select method has the following parameters:
- dataframe (pd.DataFrame): the dataframe for which to apply the selector
- target (str): the name of the column that the feature columns will be compared with
This method returns a list of the column names selected from the dataframe.
Example using KullbackLeiblerSelector
First of all, you should install kydavra if you don’t have it yet:
Now, you can import the selector:
Import a dataset and create a dataframe out of it:
df = pd.read_csv('./heart.csv')
As our selector expects numerical valued features, let’s select the columns that have a numerical data type:
Let’s instanciate our selector and choose the features compared with the ‘target’ column:
The selector returns the list of column names of the features that have a divergence with the choosen column between min_divergence and max_divergence.
With the heart.csv dataset in this example, the selector returns the following columns:
If we limit the divergence of the selected columns to be between 1 and 3 relative to the target column, we get the following:
Use case
Continuing with the heart disease dataset, let’s create a classification model that will predict wether a patient has heart disease or not. We will observe how the KullbackLeiblerSelector will improve the performance of the model.
You can find the dataset here: https://www.kaggle.com/ronitf/heart-disease-uci.
Actually we will create two models. One will be trained without feature selection and one trained with selected features from our selector.
Let’s import the dataset first:
df = pd.read_csv('./heart.csv')
X = df.drop(columns=['target'])
y = df['target']
Now let’s create the model without the selector:
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y)
clf = LogisticRegression().fit(X_train, y_train)
Now, let’s add some metrics to be able to compare the two models:
print('Without selector:')
print(f'accuracy: {accuracy_score(y_test, clf.predict(X_test)):.2f}')
print(f'recall {recall_score(y_test, clf.predict(X_test)):.2f}')
print(f'AUC {roc_auc_score(y_test, clf.predict(X_test)):.2f}')
Fine. Let’s do the same thing for the second model. But now we will apply our selector. First, import it:
We will use the columns that have a divergence between 1 and 3 relative to the target column (which in this dataset is named ‘target’ for convenience):
cols = kullback.select(df, 'target')
print(f'\nselected columns: {cols}')
Now, let’s select these columns from the DataFrame. The target column remains the same:
y = df['target']
Continue with creating the model and printing the metrics:
clf_with_selector = LogisticRegression().fit(X_train, y_train)
print('\nWith selector:')
print(f'accuracy: {accuracy_score(y_test, clf_with_selector.predict(X_test)):.2f}')
print(f'recall {recall_score(y_test, clf_with_selector.predict(X_test)):.2f}')
print(f'AUC {roc_auc_score(y_test, clf_with_selector.predict(X_test)):.2f}')
Here is what we get as an output:
accuracy: 0.82
recall 0.76
AUC 0.82
selected columns: ['sex', 'restecg', 'oldpeak']With selector:
accuracy: 0.80
recall 0.88
AUC 0.80
We observe that the accuray and AUC ROC metrics got slightly worse. Instead the recall improved a lot. In different cases the results may differ. Though, experimenting with the selector’s parameters may cause the difference in the result. Also, don’t forget to consider other factors, such as the dataset or the model used.
Made with ❤ by Sigmoid.
Follow us on Facebook, Instagram and LinkedIn:
https://www.facebook.com/sigmoidAI
https://www.instagram.com/sigmo.ai/
https://www.linkedin.com/company/sigmoid/